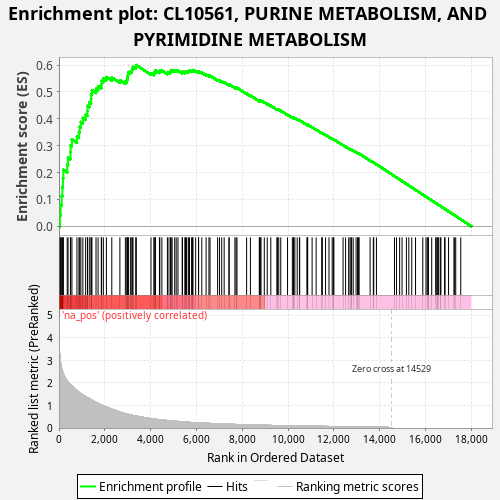
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL10561, PURINE METABOLISM, AND PYRIMIDINE METABOLISM |
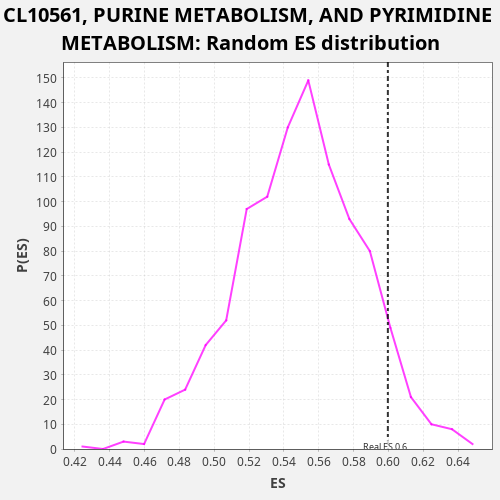
| Enrichment Score (ES) | 0.5997115 |
| Normalized Enrichment Score (NES) | 1.0917199 |
| Nominal p-value | 0.073 |
| FDR q-value | 0.6917454 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | PRPSAP2 | 39 | 3.197 | 0.0413 | Yes |
| 2 | NT5C3B | 69 | 2.917 | 0.0794 | Yes |
| 3 | ADCY9 | 116 | 2.652 | 0.1129 | Yes |
| 4 | PRKG2 | 147 | 2.556 | 0.1460 | Yes |
| 5 | NPR3 | 171 | 2.470 | 0.1784 | Yes |
| 6 | ADCY5 | 190 | 2.423 | 0.2103 | Yes |
| 7 | SLC5A2 | 359 | 2.087 | 0.2293 | Yes |
| 8 | GUCY1A2 | 394 | 2.036 | 0.2551 | Yes |
| 9 | PDE8A | 499 | 1.920 | 0.2754 | Yes |
| 10 | AKAP11 | 506 | 1.916 | 0.3012 | Yes |
| 11 | IMPDH2 | 564 | 1.867 | 0.3234 | Yes |
| 12 | ADCY4 | 783 | 1.664 | 0.3338 | Yes |
| 13 | DTYMK | 871 | 1.585 | 0.3505 | Yes |
| 14 | RGS2 | 903 | 1.559 | 0.3700 | Yes |
| 15 | RRM2B | 954 | 1.527 | 0.3880 | Yes |
| 16 | ADCY7 | 1042 | 1.467 | 0.4031 | Yes |
| 17 | CMPK2 | 1163 | 1.392 | 0.4153 | Yes |
| 18 | CANT1 | 1242 | 1.348 | 0.4293 | Yes |
| 19 | AKAP5 | 1245 | 1.347 | 0.4475 | Yes |
| 20 | AKAP1 | 1322 | 1.310 | 0.4611 | Yes |
| 21 | TK1 | 1399 | 1.259 | 0.4739 | Yes |
| 22 | PDE1A | 1404 | 1.258 | 0.4908 | Yes |
| 23 | NPR2 | 1438 | 1.236 | 0.5058 | Yes |
| 24 | PDE1C | 1616 | 1.133 | 0.5113 | Yes |
| 25 | UCK1 | 1706 | 1.096 | 0.5212 | Yes |
| 26 | NME1 | 1846 | 1.031 | 0.5275 | Yes |
| 27 | TPMT | 1859 | 1.023 | 0.5407 | Yes |
| 28 | PRKAR2A | 1936 | 0.989 | 0.5499 | Yes |
| 29 | ENTPD1 | 2069 | 0.938 | 0.5553 | Yes |
| 30 | DCTD | 2305 | 0.837 | 0.5535 | Yes |
| 31 | ENPP3 | 2658 | 0.705 | 0.5434 | Yes |
| 32 | PDE3B | 2897 | 0.637 | 0.5387 | Yes |
| 33 | AMPD3 | 2959 | 0.621 | 0.5438 | Yes |
| 34 | PDE11A | 2986 | 0.616 | 0.5507 | Yes |
| 35 | DPYD | 3000 | 0.612 | 0.5583 | Yes |
| 36 | NPR1 | 3014 | 0.608 | 0.5658 | Yes |
| 37 | AMPD1 | 3026 | 0.606 | 0.5735 | Yes |
| 38 | NT5C2 | 3116 | 0.584 | 0.5764 | Yes |
| 39 | APRT | 3181 | 0.570 | 0.5806 | Yes |
| 40 | ADCY2 | 3200 | 0.565 | 0.5873 | Yes |
| 41 | NUDT9 | 3231 | 0.556 | 0.5932 | Yes |
| 42 | CCDC102B | 3350 | 0.533 | 0.5938 | Yes |
| 43 | PDE5A | 3374 | 0.528 | 0.5997 | Yes |
| 44 | ENTPD8 | 4015 | 0.419 | 0.5695 | No |
| 45 | ENTPD4 | 4145 | 0.398 | 0.5677 | No |
| 46 | UPP1 | 4156 | 0.396 | 0.5726 | No |
| 47 | TTC5 | 4205 | 0.390 | 0.5752 | No |
| 48 | ADCY8 | 4209 | 0.389 | 0.5803 | No |
| 49 | AMPD2 | 4385 | 0.369 | 0.5755 | No |
| 50 | HDDC3 | 4402 | 0.367 | 0.5796 | No |
| 51 | GMPR2 | 4492 | 0.356 | 0.5795 | No |
| 52 | ADCY6 | 4732 | 0.331 | 0.5706 | No |
| 53 | ENTPD2 | 4758 | 0.329 | 0.5736 | No |
| 54 | AKAP7 | 4852 | 0.320 | 0.5728 | No |
| 55 | DGUOK | 4862 | 0.319 | 0.5766 | No |
| 56 | PAICS | 4897 | 0.315 | 0.5790 | No |
| 57 | PDE4C | 4938 | 0.311 | 0.5810 | No |
| 58 | NME1-NME2 | 5051 | 0.301 | 0.5788 | No |
| 59 | NME2 | 5115 | 0.296 | 0.5793 | No |
| 60 | ENPP1 | 5193 | 0.288 | 0.5789 | No |
| 61 | DUT | 5379 | 0.272 | 0.5723 | No |
| 62 | GUCY1A1 | 5391 | 0.271 | 0.5753 | No |
| 63 | NPPB | 5494 | 0.263 | 0.5732 | No |
| 64 | PRKACG | 5523 | 0.261 | 0.5752 | No |
| 65 | PRPS1L1 | 5568 | 0.259 | 0.5763 | No |
| 66 | ENTPD5 | 5643 | 0.253 | 0.5755 | No |
| 67 | UCKL1 | 5674 | 0.251 | 0.5773 | No |
| 68 | PDE9A | 5697 | 0.249 | 0.5794 | No |
| 69 | AK7 | 5788 | 0.243 | 0.5777 | No |
| 70 | ITPA | 5830 | 0.241 | 0.5787 | No |
| 71 | TK2 | 5848 | 0.240 | 0.5810 | No |
| 72 | AK3 | 5966 | 0.233 | 0.5776 | No |
| 73 | IMPDH1 | 6091 | 0.227 | 0.5737 | No |
| 74 | ADA2 | 6095 | 0.226 | 0.5766 | No |
| 75 | TYMP | 6234 | 0.219 | 0.5719 | No |
| 76 | NT5C3A | 6422 | 0.209 | 0.5643 | No |
| 77 | IRAG1 | 6546 | 0.203 | 0.5601 | No |
| 78 | RAPGEF3 | 6593 | 0.201 | 0.5603 | No |
| 79 | PDE3A | 6924 | 0.186 | 0.5443 | No |
| 80 | AK6 | 7007 | 0.182 | 0.5422 | No |
| 81 | NPPA | 7103 | 0.178 | 0.5393 | No |
| 82 | CMPK1 | 7210 | 0.174 | 0.5357 | No |
| 83 | ADCY3 | 7421 | 0.166 | 0.5262 | No |
| 84 | AKAP8 | 7425 | 0.166 | 0.5283 | No |
| 85 | AK2 | 7679 | 0.158 | 0.5163 | No |
| 86 | PNP | 7730 | 0.156 | 0.5156 | No |
| 87 | AK8 | 7769 | 0.155 | 0.5156 | No |
| 88 | ENTPD6 | 8191 | 0.141 | 0.4939 | No |
| 89 | ADK | 8356 | 0.136 | 0.4866 | No |
| 90 | ADSL | 8743 | 0.126 | 0.4666 | No |
| 91 | PRKACB | 8761 | 0.125 | 0.4674 | No |
| 92 | ARMC5 | 8808 | 0.124 | 0.4665 | No |
| 93 | DCTPP1 | 8813 | 0.124 | 0.4680 | No |
| 94 | NT5E | 8962 | 0.120 | 0.4613 | No |
| 95 | NME4 | 9090 | 0.117 | 0.4558 | No |
| 96 | JMY | 9247 | 0.112 | 0.4486 | No |
| 97 | HDHD5 | 9512 | 0.107 | 0.4352 | No |
| 98 | GUCY1B1 | 9535 | 0.107 | 0.4354 | No |
| 99 | AKAP8L | 9593 | 0.105 | 0.4337 | No |
| 100 | NT5C1B-RDH14 | 9681 | 0.104 | 0.4302 | No |
| 101 | PRPSAP1 | 9977 | 0.097 | 0.4150 | No |
| 102 | PDE7A | 10188 | 0.094 | 0.4045 | No |
| 103 | CLCN2 | 10239 | 0.093 | 0.4030 | No |
| 104 | NT5C1A | 10245 | 0.093 | 0.4039 | No |
| 105 | AK9 | 10296 | 0.092 | 0.4024 | No |
| 106 | NT5C1B | 10399 | 0.090 | 0.3979 | No |
| 107 | ATP2B3 | 10497 | 0.088 | 0.3936 | No |
| 108 | ABHD15 | 10507 | 0.088 | 0.3943 | No |
| 109 | NME6 | 10823 | 0.082 | 0.3778 | No |
| 110 | NME3 | 10848 | 0.082 | 0.3776 | No |
| 111 | CDA | 10857 | 0.082 | 0.3782 | No |
| 112 | PDE2A | 11050 | 0.078 | 0.3685 | No |
| 113 | SLFN12 | 11225 | 0.076 | 0.3598 | No |
| 114 | PDE4DIP | 11469 | 0.072 | 0.3472 | No |
| 115 | PALM2AKAP2 | 11496 | 0.072 | 0.3467 | No |
| 116 | UMPS | 11637 | 0.069 | 0.3398 | No |
| 117 | PDE8B | 11787 | 0.067 | 0.3324 | No |
| 118 | NUDT5 | 11927 | 0.065 | 0.3254 | No |
| 119 | SLC6A2 | 11976 | 0.064 | 0.3236 | No |
| 120 | PDE10A | 12001 | 0.064 | 0.3231 | No |
| 121 | AK5 | 12401 | 0.059 | 0.3016 | No |
| 122 | ADSS2 | 12511 | 0.057 | 0.2962 | No |
| 123 | AK4 | 12646 | 0.055 | 0.2895 | No |
| 124 | OSTN | 12706 | 0.054 | 0.2869 | No |
| 125 | GNAL | 12749 | 0.054 | 0.2853 | No |
| 126 | AK1 | 12775 | 0.053 | 0.2846 | No |
| 127 | NTPCR | 12844 | 0.052 | 0.2815 | No |
| 128 | PDE4A | 12969 | 0.051 | 0.2753 | No |
| 129 | RRM2 | 13036 | 0.050 | 0.2722 | No |
| 130 | PPAT | 13048 | 0.050 | 0.2723 | No |
| 131 | PHYHIPL | 13081 | 0.049 | 0.2712 | No |
| 132 | PRKAR1B | 13100 | 0.049 | 0.2708 | No |
| 133 | AKAP10 | 13581 | 0.042 | 0.2445 | No |
| 134 | PRKAR2B | 13722 | 0.041 | 0.2372 | No |
| 135 | PDE1B | 13741 | 0.040 | 0.2367 | No |
| 136 | ADCY1 | 13858 | 0.039 | 0.2308 | No |
| 137 | CTPS2 | 14646 | -0.000 | 0.1867 | No |
| 138 | PRKACA | 14736 | -0.000 | 0.1817 | No |
| 139 | UPRT | 14871 | -0.000 | 0.1742 | No |
| 140 | PRPS2 | 14976 | -0.000 | 0.1683 | No |
| 141 | PRKAR1A | 15166 | -0.000 | 0.1577 | No |
| 142 | PDE4D | 15272 | -0.000 | 0.1518 | No |
| 143 | GDA | 15400 | -0.000 | 0.1447 | No |
| 144 | NT5C | 15565 | -0.000 | 0.1355 | No |
| 145 | GMPR | 15882 | -0.000 | 0.1178 | No |
| 146 | UCK2 | 16021 | -0.000 | 0.1101 | No |
| 147 | CORIN | 16076 | -0.000 | 0.1071 | No |
| 148 | AKAP4 | 16111 | -0.000 | 0.1052 | No |
| 149 | PRPS1 | 16125 | -0.000 | 0.1044 | No |
| 150 | DCK | 16271 | -0.000 | 0.0963 | No |
| 151 | NPPC | 16446 | -0.000 | 0.0865 | No |
| 152 | GMPS | 16470 | -0.000 | 0.0853 | No |
| 153 | ALDH7A1 | 16537 | -0.000 | 0.0816 | No |
| 154 | NUDT2 | 16541 | -0.000 | 0.0814 | No |
| 155 | HPRT1 | 16577 | -0.000 | 0.0794 | No |
| 156 | RRM1 | 16650 | -0.000 | 0.0754 | No |
| 157 | ENTPD3 | 16672 | -0.000 | 0.0742 | No |
| 158 | PDE7B | 16833 | -0.000 | 0.0652 | No |
| 159 | CTPS1 | 16853 | -0.000 | 0.0642 | No |
| 160 | GNG7 | 17006 | -0.000 | 0.0557 | No |
| 161 | PDE4B | 17242 | -0.000 | 0.0425 | No |
| 162 | PRKG1 | 17273 | -0.000 | 0.0408 | No |
| 163 | AKAP14 | 17320 | -0.000 | 0.0382 | No |
| 164 | QRICH1 | 17539 | -0.000 | 0.0260 | No |